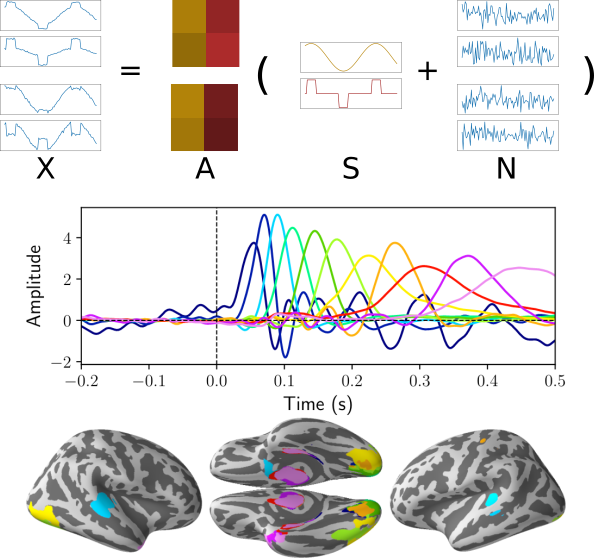
MultiView ICA¶
Code accompanying the paper MultiViewICA https://arxiv.org/pdf/2006.06635.pdf
Documentation: https://hugorichard.github.io/multiviewica/
Install¶
Clone the repository
git clone https://github.com/hugorichard/multiviewica.git
Move into the multiviewica directory
cd multiviewica
Install MultiView ICA
pip install -e .
Requirements¶
For the core algorithms: * numpy >= 1.16 * scipy >= 1.12 *
scikit-learn >= 0.20 * python-picard >= 0.4
(pip install python-picard)
For the Experiments: * nibabel (>=2.3.3) * mne (>=0.20) * nilearn
(>=0.5) * fastsrm (pip install fastsrm)
Experiments¶
Synthetic experiment¶
Move into the multiviewica directory
cd multiviewica
Run the experiment on synthetic data
python examples/synthetic_experiment.py
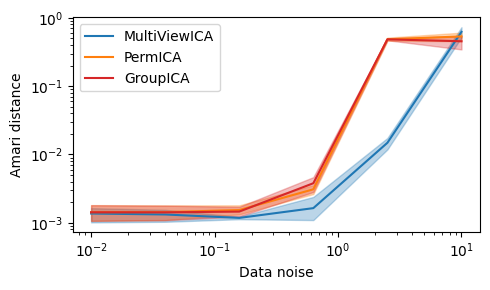
Synthetic Experiment¶
In order to reproduce the figure in the paper, use (might take a long time):
# sigmas: data noise
# m: number of subjects
# k: number of components
# n: number of samples
sigmas = np.logspace(-2, 1, 21)
n_seeds = 100
m, k, n = 10, 15, 1000
Experiments on fMRI data¶
Download and mask Sherlock data¶
Move into the data directory
cd multiviewica/data
Launch the download script (Runtime 34m6.751s)
bash download_data.sh
Mask the data (Runtime 15m27.104s)
python mask_data.py
Reconstructing BOLD signal of missing subjects¶
Move into the real_data_experiments directory
cd multiviewica/real_data_experiments
Run the experiment on masked data (Runtime 30m55.347s)
python reconstruction_experiment.py
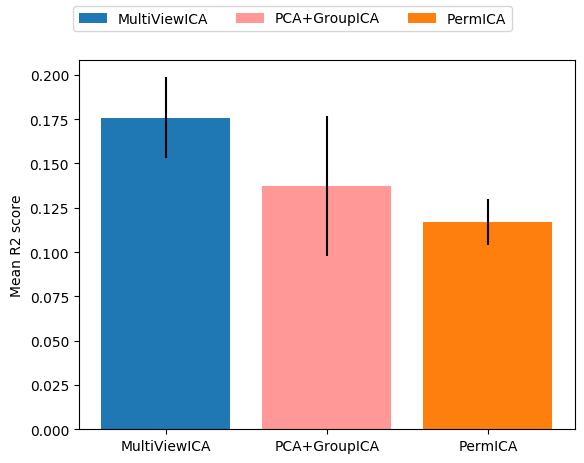
Reconstruction experiment¶
This runs the experiment with n_components = 5 and benchmark
PCA + GroupICA, PermICA and MultiView ICA with subject
specific PCA for dimension reduction in PCA + GroupICA and SRM for
PermICA and MultiView ICA.
Timesegment matching¶
Move into the real_data_experiment directory
cd multiviewica/real_data_experiments
Run the experiment on masked data (Runtime 17m39.520s)
python timesegment_matching.py
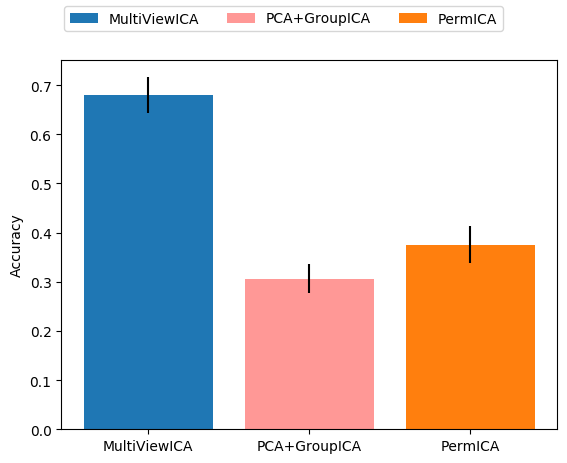
Timesegment matching¶
This runs the experiment with n_components = 5 and benchmark
PCA + GroupICA, PermICA and MultiView ICA with subject
specific PCA for dimension reduction in PCA + GroupICA and SRM for
PermICA and MultiView ICA.
Cite¶
If you use this code in your project, please cite:
@inproceedings{NEURIPS2020_de03beff,
author = {Richard, Hugo and Gresele, Luigi and Hyvarinen, Aapo and Thirion, Bertrand and Gramfort, Alexandre and Ablin, Pierre},
booktitle = {Advances in Neural Information Processing Systems},
editor = {H. Larochelle and M. Ranzato and R. Hadsell and M. F. Balcan and H. Lin},
pages = {19149--19162},
publisher = {Curran Associates, Inc.},
title = {Modeling Shared responses in Neuroimaging Studies through MultiView ICA},
url = {https://proceedings.neurips.cc/paper/2020/file/de03beffeed9da5f3639a621bcab5dd4-Paper.pdf},
volume = {33},
year = {2020}
}